For Researchers
Use our data
How to use this site
You can filter and search content by key words. Once you find a project you are interested in pursing, select the contact button to access our contact information and to indicate you're working on the idea.
Project types
This site contains ideas, questions, observations that our scientists have come up with in the process of collecting data. There are more ideas than we can ever pursue ourselves, and our hope is other researchers will be able to take these ideas and make new discoveries.
Most recent additions
- ### Overview This dataset contains simulated data of actin filaments using the monomer-scale model [ReaDDy](https://github.com/simularium/readdy-models) and fiber-scale model [Cytosim](https://github.com/simularium/Cytosim). Simulation conditions test compression of a single 500 nm actin filament at different velocities, as well as control simulations with no compression. Compression simulations use the prefix `ACTIN_COMPRESSION_VELOCITY`; control simulations use the prefix `ACTIN_NO_COMPRESSION`. Compression simulation keys include the following: - `0047` = 4.7 μm/s - `0150` = 15 μm/s - `0470` = 47 μm/s - `1500` = 150 μm/s For _ReaDDy_, the actin filament is composed of particles, each representing an actin monomer, initialized based on measurements from an actin crystal structure. Compression was implemented by linearly displacing the three monomers at the pointed end for one time step and then allowing the structure to relax for nine time steps, until the fiber was compressed to 350 nm. Each simulation condition contains 5 replicates. For _Cytosim_, the actin filament is modeled as a 500 nm fiber anchored by two fiber-binding linkers spaced 10 nm apart at each end. The linkers on one of the ends were translated linearly for one time step and allowed to relax for nine time steps, until the fiber was compressed to 350 nm. Each simulation condition contains 5 replicates, with replicates corresponding to random seeds 1, 2, 3, 4, and 5. The dataset has the following structure: ``` . ├── fiber_scale_cytosim │ ├── ACTIN_COMPRESSION_VELOCITY │ └── ACTIN_NO_COMPRESSION ├── monomer_scale_readdy │ ├── ACTIN_COMPRESSION_VELOCITY │ └── ACTIN_NO_COMPRESSION ├── actin_compression_aligned_fibers.json ├── actin_compression_combined_metrics.csv ├── actin_compression_pca_results.csv ├── actin_compression_pca_trajectories.json ├── actin_compression_pca_transforms.json ├── actin_compression.simularium └── README.md ``` - `fiber_scale_cytosim` = simulation, analysis, and visualization data for fiber-scale model Cytosim - `monomer_scale_readdy` = simulation, analysis, and visualization data for monomer-scale model ReaDDy - `actin_compression_aligned_fibers.json` = positions of aligned fibers for both models - `actin_compression_combined_metrics.csv` = compression metrics calculated on both models - `actin_compression_pca_results.csv` = PCA embedding on filament from both models - `actin_compression_pca_trajectories.json` = trajectories in PCA space for both models - `actin_compression_pca_transforms.json` = PCA inverse transformed filament shapes - `actin_compression.simularium` = Simularium visualization of both models For monomer-scale _ReaDDy_, the dataset has the following structure. Note that `(index)` indicates the index of job in the batch job array, while `(repeat)` indicates the replicate number. ``` . └── monomer_scale_readdy ├── ACTIN_COMPRESSION_VELOCITY │ ├── analysis │ │ ├── ACTIN_COMPRESSION_VELOCITY_all_samples_aligned.csv │ │ ├── ACTIN_COMPRESSION_VELOCITY_all_samples_unaligned.csv │ │ └── ACTIN_COMPRESSION_VELOCITY_compression_metrics.csv │ ├── data │ │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(repeat).pkl │ ├── outputs │ │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(index).h5 │ ├── parameters │ │ └── ACTIN_COMPRESSION_VELOCITY.xlsx │ ├── samples │ │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(repeat).csv │ └── viz │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(repeat).simularium └── NO_COMPRESSION ├── analysis │ └── ACTIN_NO_COMPRESSION_compression_metrics.csv ├── data │ └── ACTIN_NO_COMPRESSION_(repeat).pkl ├── outputs │ └── ACTIN_NO_COMPRESSION_(index).h5 ├── parameters │ └── ACTIN_NO_COMPRESSION.xlsx ├── samples │ └── ACTIN_NO_COMPRESSION_(repeat).csv └── viz └── ACTIN_NO_COMPRESSION_(repeat).simularium ``` - `analysis` = various analysis files - `data` = simulation outputs parsed into data structures - _Note that these `.pkl` files containing the parsed data are provided for convenience. You may encounter errors when unpickling them due to dependency conflicts. You can safely delete these files and allow the pipeline to regenerate them directly from the raw `.h5` simulation outputs in the `output` folder._ - `outputs` = raw simulation outputs - `parameters` = simulation parameters - `samples` = simulation outputs sampled to 200 filament points x 201 time points - `viz` = visualization files For fiber-scale _Cytosim_, the dataset has the following structure. Note that `(index)` indicates the index of job in the batch job array, while `(seed)` indicates the random seed used. ``` . └── fiber_scale_cytosim ├── ACTIN_COMPRESSION_VELOCITY │ ├── analysis │ │ ├── ACTIN_COMPRESSION_VELOCITY_all_samples_aligned.csv │ │ ├── ACTIN_COMPRESSION_VELOCITY_all_samples_unaligned.csv │ │ └── ACTIN_COMPRESSION_VELOCITY_compression_metrics.csv │ ├── configs │ │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(index).cym │ ├── data │ │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(seed).csv │ ├── outputs │ │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(index)/ │ ├── samples │ │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(seed).csv │ └── viz │ └── ACTIN_COMPRESSION_VELOCITY_(velocity)_(seed).simularium └── ACTIN_NO_COMPRESSION ├── analysis │ └── ACTIN_NO_COMPRESSION_compression_metrics.csv ├── configs │ └── ACTIN_NO_COMPRESSION_(index).cym ├── data │ └── ACTIN_NO_COMPRESSION_(seed).csv ├── outputs │ └── ACTIN_NO_COMPRESSION_(index)/ ├── samples │ └── ACTIN_NO_COMPRESSION_(seed).csv └── viz └── ACTIN_NO_COMPRESSION_(seed).simularium``` - `analysis` = various analysis files - `configs` = model configuration files - `data` = simulation outputs formatted into tidy data format - `outputs` = raw simulation outputs - `samples` = simulation outputs sampled to 200 filament points x 201 time points - `viz` = visualization files ### Usage For documentation on how to use and interact with this dataset please refer to [https://docs.quiltdata.com/walkthrough/getting-data-from-a-package](https://docs.quiltdata.com/walkthrough/getting-data-from-a-package). ### License For questions on licensing please refer to [https://www.allencell.org/terms-of-use.html](https://www.allencell.org/terms-of-use.html).
- 2
- 2
- actin
- computational modeling
- 2
- 2
- actin
- computational modeling
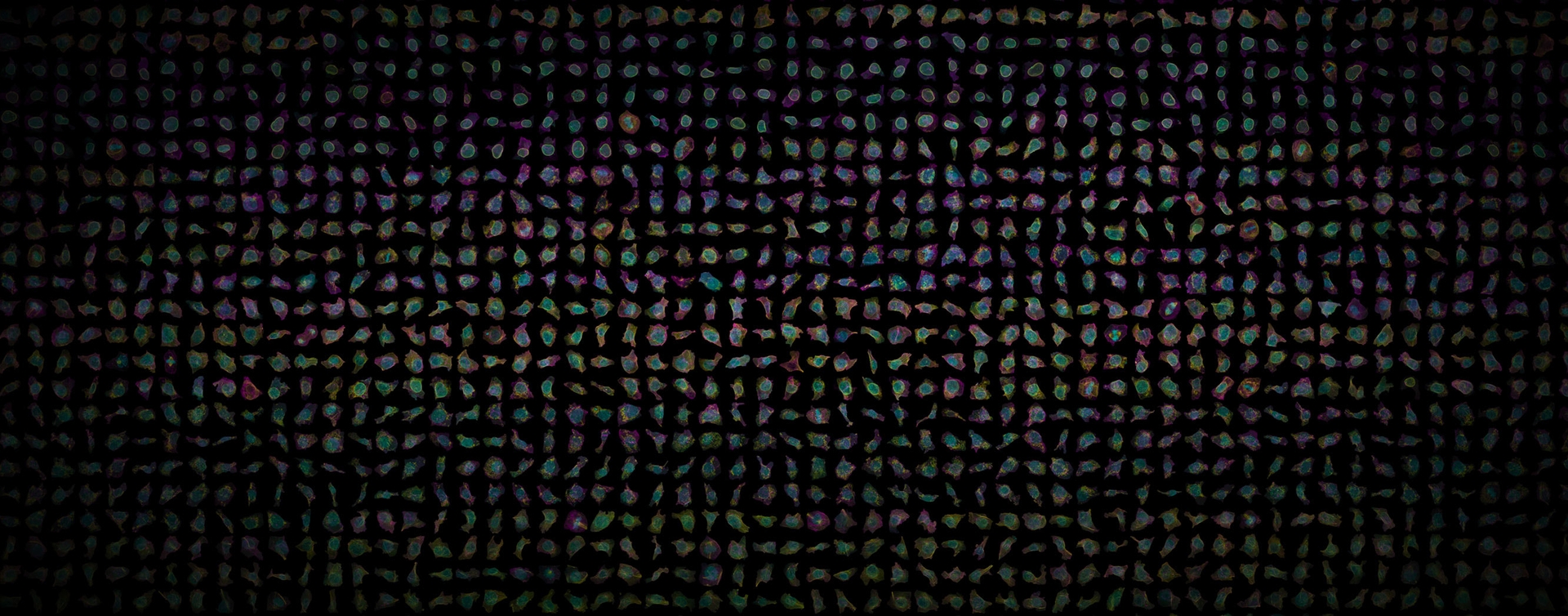
- Description for released EMT dataset in the markdown file for the dataset.
- 2
- 2
- EMT
- migration